Processing Large Files in Python
If any of you are followers of Tech Twitter, you'll know that the 1 billion row challenge was something that was doing the rounds a couple of weeks ago in the Java community. Since then, it's been an excuse for people to use other languages to tackle the problem of quickly and efficiently solving a simple problem of parsing a very large file and performing some simple operations on the contents of it.
Processing a file is a straightforward endevour. You open the file, read the contents and then do something with it. The problem with this is that it's not very efficient.
Simple approach (not optimal for large files)
The simplest approach to processing a file is to read it line by line and perform the operations on each line. Below is a simple example of this:
def simple_get_weather_data(filepath: str) -> Dict[str, List[float]]:
results = {}
with open(filepath, "r") as file:
for line in file:
location, measurement = line.split(';')
if results.get(location):
results[location].append(float(measurement))
else:
results[location] = [float(measurement)]
return results
On my 2021 Macbook Pro, this takes around 0.2 seconds to process a file with 100k rows. This file is around 1.5MB in size. If we have a file that is 10M rows and 152MB, this takes around 3.5 seconds to process. When we scale to a 1B row file, this is an astonishing 15GB in size, and takes around 7 minutes to process. This is not ideal.
Why is this so slow?
It's important when thinking of optimising a solution to understand what the bottlenecks are. On MacOS, we can take a look at Activity Monitor to analyse the CPU usage, Memory usage and Disk Usage. When we run the above code, we can see that the CPU usage is around 100% on one process and the memory usage is growing steadily with gigabytes of data being read into memory. This is all while disk usage is relatively low.
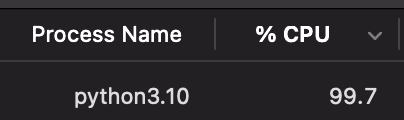
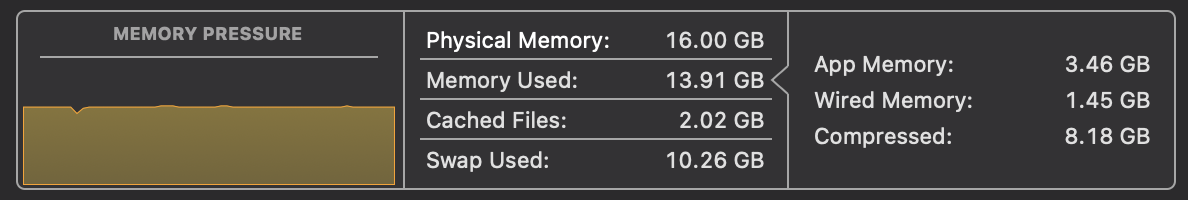
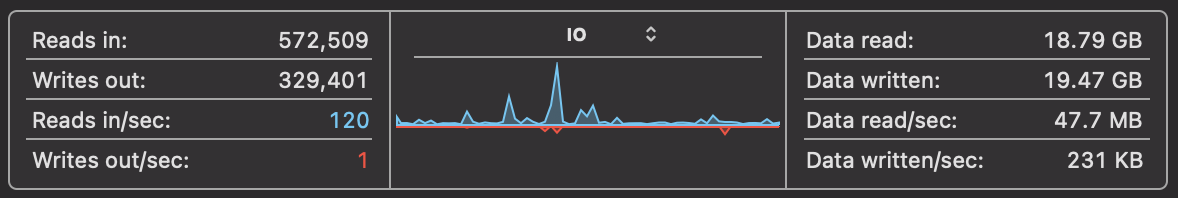
We need to address this situation. If we can spread the CPU work onto multiple cores, reduce memory pressure and increase the disk usage, we can speed up the processing of the file.
Using multiprocessing
I have a 10 core processor on my MacBook Pro, so I can use multiprocessing to spread the work across multiple cores. Let's say we want to split the file in such a way that each core can process a part of the file, and we'll want 10 file chunks to process.
First, we'll need to split the file into 10 chunks.
The python files API provides us with a few functions to help us achieve
this. We can use the seek function to move the file pointer to a specific
location in the file. We need to ensure we don't just find an arbitrary location
in the file, we need to find a line break, so we use the readline function to
read to the end of the line, then we can use the tell function to
find the current location of the file pointer.
def get_file_chunks(file_name: str, cpu_cores: int):
file_size = os.path.getsize(file_name)
chunk_size = file_size // cpu_cores
chunks = []
with open(file_name, "r") as file:
chunk_start = 0
while chunk_start < file_size:
chunk_end = min(file_size, chunk_start + chunk_size)
file.seek(chunk_end)
file.readline()
chunk_end = file.tell()
chunks.append((file_name, chunk_start, chunk_end))
chunk_start = chunk_end
return chunks
This gives us a list of 10 chunks of the file from which we can process. We can
then use the multiprocessing library to process each chunk in parallel.
def process_file(cpu_count: int, chunks: list):
with mp.Pool(cpu_count) as p:
# Run chunks in parallel
chunk_results = p.starmap(
_process_file_chunk,
chunks,
)
# Combine results
results = {}
for chunk_result in chunk_results:
for location, measurements in chunk_result.items():
if location not in results:
results[location] = measurements
else:
results[location].extend(measurements)
return results
def _process_file_chunk(file_name: str, start: int, end: int):
results = {}
with open(file_name, "r") as file:
file.seek(start)
for line in file:
start += len(line)
if start > end:
break
location, measurement = line.split(';')
if results.get(location):
results[location].append(float(measurement))
else:
results[location] = [float(measurement)]
return results
This works incredibly well for medium sized files, as this allows us to utilise all of the CPU cores available to us. However, when we scale up to a 1B row file, we run into some issues. The first issue is that we run out of memory.
A recap on floats in Python
In python, floats are objects. This means that they have a lot of overhead associated with them, taking up a lot of memory. This is because the number is stored as an 8 byte double, and the overhead of the object is 16 bytes. This means that if we have a list of 1B floats, we're storing 24 bytes per float, which is 24GB of memory.
This is very different to a language like C. In C, a double is stored as a 8 byte double. This means that a list of 1B doubles is 8GB of memory. This is a lot less than the 24GB we're using in Python.
So what can we do?
In our example, we're storing the measurements for each location in a list. This means that we're storing a list of 1B floats in memory. If instead, we could calculate the mean, min and max as we go, we no longer need to store the list of measurements in memory. This will dramatically reduce the memory footprint of our process.
There are around 45k cities in our dataset, so if we adopt the above approach, we'll be storing 45,000 (cities) x 4 (min, max, count & sum) x 24 bytes of memory, which is 4.3MB. This is a lot less than the 52GB we were using before.
def process_file(cpu_count: int, chunks: list):
with mp.Pool(cpu_count) as p:
# Run chunks in parallel
chunk_results = p.starmap(
_process_file_chunk,
chunks,
)
# Combine results
results = {} # {location: [min, max, sum, count]}
for chunk_result in chunk_results:
for location, measurements in chunk_result.items():
if location not in results:
results[location] = measurements
else:
if measurements[0] < results[location][0]:
results[location][0] = measurements[0]
if measurements[1] > results[location][1]:
results[location][1] = measurements[1]
results[location][2] += measurements[2]
results[location][3] += measurements[3]
return results
def _process_file_chunk(file_name: str, start: int, end: int) -> dict:
results = {} # {location: [min, max, sum, count]}
with open(file_name, "r") as file:
file.seek(start)
for line in file:
start += len(line)
if start > end:
break
location, measurement = line.split(';')
measurement = float(measurement)
if results.get(location):
# If less than min, replace min
if measurement < results[location][0]:
results[location][0] = measurement
# If more than max, replace max
if measurement > results[location][1]:
results[location][1] = measurement
# Add to sum
results[location][2] += measurement
# Add to count
results[location][3] += 1
else:
results[location] = [measurement, measurement, measurement, 1]
return results
Now we're finally in a position where we can process a 1B row file in a reasonable amount of time. On my 2021 MacBook Pro, this takes around 2 minutes to process. This is all while having a negligible memory footprint and having multiple cores running at high CPU usage.
The cores weren't pegged at 100% CPU usage, but they were all running at around 80% CPU usage, and there were no bottlenecks in terms of IO performance or memory usage. I think that there is still some room for improvement here, but I'm not sure how much more we can squeeze out of this. If we can manage to up the CPU usage to 100%, we might be able to make this process ever so slightly faster.
We were helped by the fact that we were able to discard the list of measurements for each location, as the statistics we had to calculate were the min, max, mean (sum/count). If we had to calculate something like the median, we would have to store the list of measurements in memory, which would mean we would run out of memory.
If you have to do that, look into either using a different programming language which stores floats more efficiently. Let me know if you have any ideas on how to improve this further, I'd love to hear them!